We take single molecule approaches to important biological/biomedical problems. From real-time assessment of the dynamics of individual biomolecules or real-time tracing of dynamic processes in the cell, we can reveal the following kinds of information, which are often missed by conventional ensemble-averaging methods.
- Non-synchronizable dynamics of biomolecules
- Hidden intermediates and their kinetics
- Molecular/cellular heterogeneity
- Causal relation between mechanistically associated events
Many of the greatest challenges in molecular and cellular biology lie in the nucleus. Extremely compact and hierarchical packaging of meters-long chromosomes and highly crowded nuclear environment make the direct assessment of chromatin arrangement and the observation of dynamic nuclear events difficult and also make these behaviors diverge from what is predicted from in vitro systems. We focus on the following themes central to the dynamic processes in nucleus from biophysical perspectives using single molecule and novel imaging techniques.
Dynamics of Molecular Motors
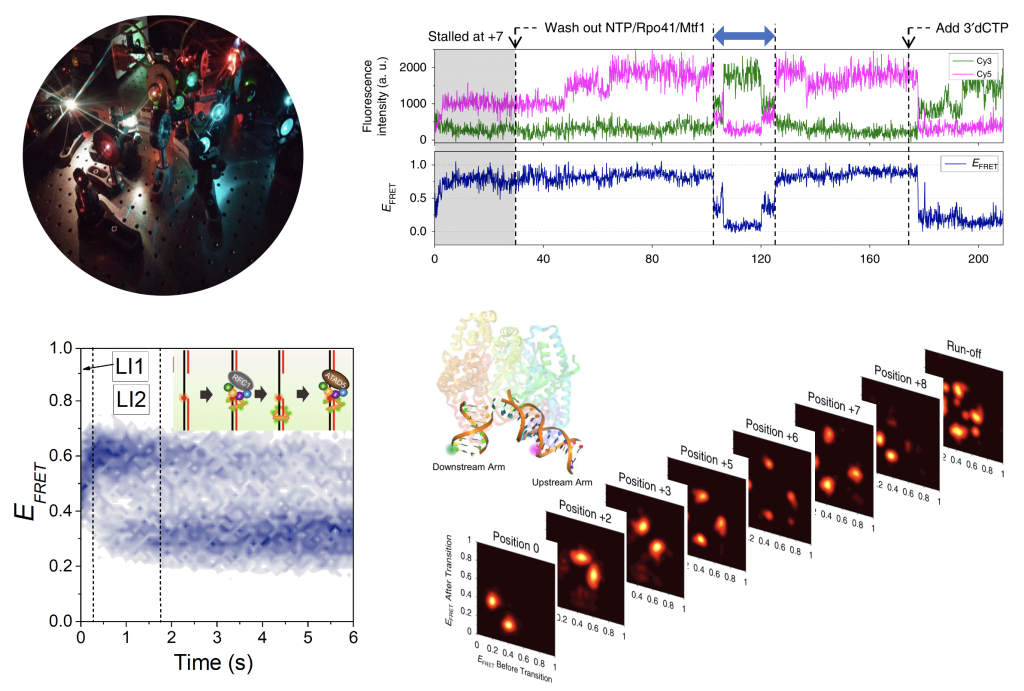
We investigate the working principles of molecular machineries that assemble on and interact with nucleic acids to perform functions in the maintenance, replication, and transcription of genome. From single molecule fluorescence measurements and force manipulation, we aim to build mechanistic models of these nucleic acids molecular motors. Conformational transitions of the molecular machineries during their interaction with nucleic acids are keys to understanding how they achieve high target specificity and processivity, as well as how their functions are regulated. Single molecule approaches can illuminate this aspect by directly recording the motions of motor proteins along nucleic acids, revealing reversible or irreversible conformational transitions. We aim to dissect the molecular details of such conformational dynamics during (1) replication by studying the interplays of DNA polymerases, PCNA, and helicases with DNA and (2) transcription initiation by mitochondrial transcription system.
4D Chromatin Dynamics
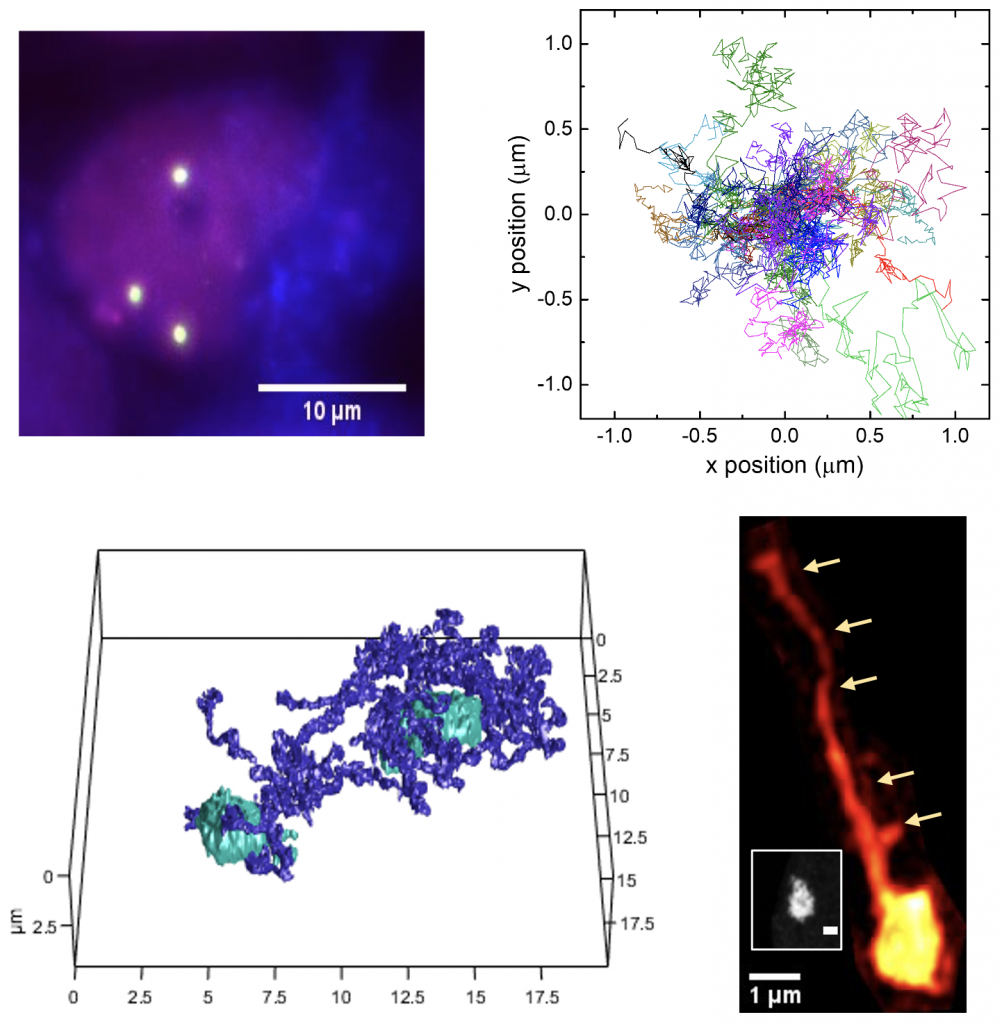
We are interested in uncovering the molecular level principles that govern the spatiotemporal organization of chromatin and its epigenetic control for regulating gene expression and eventually determining the fate of the cell. We take novel live imaging techniques to visualize the molecular-scale dynamics occurring within the nucleus. The chromosomes are known to undergo complex processes of rearrangement to perform their functions in genome maintenance, gene expression, proliferation, and differentiation. Dissecting the molecular mechanisms of the chromatin reorganization thus is the key to discovering the principles of these processes. We take combined approaches of “bottom-up” single molecule experiments and “top-down” visualization of live genome to address this problem. We aim to construct the molecular level foundation to test and explain the biophysical principles governing chromatin reorganization and dynamics.
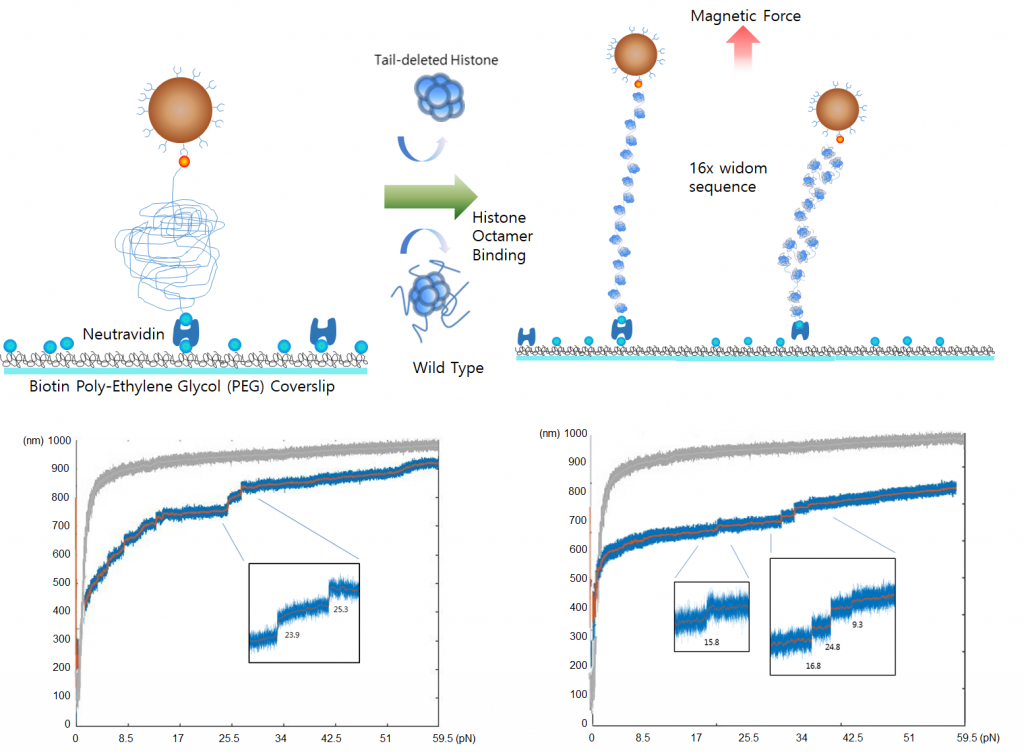
Single-Molecule Nucleosome Dynamics
We want to deeply understand the dynamical features of chromatin fibers from the simplest form of nucleosome to even higher dimensional structures. Monitoring the dynamics of single chromatin fiber gives clue to understand how the epigenetic regulation works on chromatin and thereby to the structure and function of the genetic activity.