Scientific Papers
- [42]
Eui-Jin Park and Hajin Kim
- "Live genome imaging by CRISPR engineering: progress and problems"
- Experimental & Molecular Medicine
- / 202507
- [41]
Yubin Sung, Chanwoo Kim, Hajin Kim, Kei-ichi Takata
- "Measurement of DNA Polymerase and Microhomology-Mediated End-Joining Activities by Gel Electrophoresis and smFRET Techniques"
- DNA Damage Detection (book chapter)
- / 2025
- [40]
Sihyeong Nho, Hajin Kim
- "Dynamics of nucleosomes and chromatin fibers revealed by single-molecule measurements"
- BMB Reports
- / 2025
- [39]
Sungho Kim*, Hwi Hyun*, Jae-Kyeong Im, Min Seok Lee, Hwasoo Koh, Donghoon Kang, Si-Hyeong Nho, Joo H. Kang+, Taejoon Kwon+, Hajin Kim+
- "Fast and accurate multi-bacterial identification using cleavable and FRET-based peptide nucleic acid probes"
- Biosensors and Bioelectronics
- / 2024
- [38]
Hyun Park, Go Woon Shin, Sang Min Lee, Gyu Won Jeong, Hui Young Kim, Hajin Kim, Hyun Woo Choi, Whaseon Lee-Kwon, Hyug Moo Kwon+
- "One-hit kill: On the inactivation of RNA viruses by ultraviolet (UV)-C-induced genomic damage"
- Journal of Photochemistry and Photobiology B: Biology
- / 2024
- [37]
Son Truong Le, Seoyun Choi, Seung-Won Lee, Hajin Kim, Byungchan Ahn
- "ssDNA reeling is an intermediate step in the reiterative DNA unwinding activity of the WRN-1 helicase"
- Journal of Biological Chemistry
- / 2023
- [36]
- "DNA polymerase θ-mediated repair of high LET radiation-induced complex DNA double-strand breaks"
- Nucleic Acids Research
- / 2023
- [35]
Min Seok Lee*, Hwi Hyun*, Inwon Park*, Sungho Kim*, Dong-Hyun Jang, Seonghye Kim, Jae-Kyeong Im, Hajin Kim+, Jae Hyuk Lee+, Taejoon Kwon+, Joo H. Kang+
- "Quantitative Fluorescence In Situ Hybridization (FISH) of Magnetically Confined Bacteria Enables Early Detection of Human Bacteremia"
- Small Methods
- / 2022
- [34]
Hwi Hyun*, Min Seok Lee*, Inwon Park*, Hwa Soo Ko, Seongmin Yun, Dong-Hyun Jang, Seonghye Kim, Hajin Kim, Joo H. Kang+, Jae Hyuk Lee+, Taejoon Kwon+
- "Analysis of Porcine Model of Fecal-Induced Peritonitis Reveals the Tropism of Blood Microbiome"
- Frontiers in Cellular and Infection Microbiology
- / 2021
- [33]
Narendra Chaudhary, Jae-Kyeong Im, Si-Hyeong Nho, Hajin Kim
- "Visualizing Live Chromatin Dynamics through CRISPR-Based Imaging Techniques"
- Molecules and Cells
- / 2021
- [32]
Narendra Chaudhary, Si-Hyeong Nho, Hayoon Cho, Narangerel Gantumur, Jae Sun Ra, Kyungjae Myung, Hajin Kim
- "Background-Suppressed Live Visualization of Genomic Loci with an Improved CRISPR System Based on a Split Fluorophore"
- Genome Research
- / 2020
- [31]
Byeong-Kwon Sohn*, Urmimala Basu*, Seung-Won Lee, Hayoon Cho, Jiayu Shen, Aishwarya Deshpande, Laura C. Johnson, Kalyan Das, Smita S. Patel+, Hajin Kim+
- "The Dynamic Landscape of Transcription Initiation in Yeast Mitochondria"
- Nature Communications
- / 2020
- [30]
Jun Ho Lee, Jae Hee Suh, Hyun Je Kang, Soo Youn Choi, Seok Won Jung, Whaseon Lee-Kwon, Soo-Ah Park, Hajin Kim, Byeong Jin Ye, Eun Jin Yoo, Gyu Won Jeong, Neung Hwa Park, Hyug Moo Kwon
- "Tonicity-Responsive Enhancer-Binding Protein Promotes Stemness of Liver Cancer and Cisplatin Resistance"
- EBioMedicine
- / 2020
- [29]
Urmimala Basu*, Seung-Won Lee*, Aishwarya Deshpande*, Jiayu Shen, Byeong-Kwon Sohn, Hayoon Cho, Hajin Kim+, and Smita S. Patel+
- "The C-terminal Tail of the Yeast Mitochondrial Transcription Factor Mtf1 Coordinates Template Strand Alignment, DNA Scrunching, and Timely Transition into Elongation"
- Nucleic Acids Research
- / 2020
- [28]
Xianan Qin, Lei Liu, Sang Kwon Lee, Adolfo Alsina, Teng Liu, Chao Wu, Hojeong Park, Chenglong Yu, Hajin Kim, Jun Chu, Antoine Triller, Ben Zhong Tang, Changbong Hyeon, Chan Young Park, Hyokeun Park
- "Increased Confinement and Polydispersity of STIM1 and Orai1 after Ca2+ Store Depletion"
- Biophysical Journal
- / 2020
- [27]
Seoyun Choi, Seung-Won Lee, Hajin Kim+, Byungchan Ahn+
- "Molecular Characteristics of Reiterative DNA Unwinding by the Caenorhabditis elegans RecQ Helicase"
- Nucleic Acids Research
- / 2019
- [26]
Mi-Sun Kang*, Eunjin Ryu*, Seung-Won Lee*, Jieun Park, Na Young Ha, Jae Sun Ra, Yeong Jae Kim, Jinwoo Kim, Mohamed Abdel-Rahman, Su Hyung Park, Kyoo-young Lee, Hajin Kim+, Sukhyun Kang+, Kyungjae Myung+
- "Regulation of PCNA Cycling on Replicating DNA by RFC and RFC-like Complexes"
- Nature Communications
- / 2019
- [25]
Jyoti Bhardwaj, Narendra Chaudhary, Hajin Kim, Jaesung Jang
- "Subtyping of Influenza A H1N1 Virus Using a Label-Free Electrochemical Biosensor Based on the DNA Aptamer Targeting the Stem Region of HA Protein"
- Analytica Chimica Acta
- / 2019
- [24]
Hyunju Kang*, Jejoong Yoo*, Byeong-Kwon Sohn, Seung-Won Lee, Hong Soo Lee, Wenjie Ma, Jung-Min Kee, Aleksei Aksimentiev+, Hajin Kim+
- "Sequence-Dependent DNA Condensation as a Driving Force of DNA Phase Separation"
- Nucleic Acids Research
- / 2018
- [23]
Sanjaya C. Abeysirigunawardena*, Hajin Kim*, Jonathan Lai, Kaushik Ragunathan, Mollie C. Rappé, Zaida Luthey-Schulten, Taekjip Ha+, Sarah A. Woodson+
- "Evolution of Protein-Coupled RNA Dynamics during Hierarchical Assembly of Ribosomal Complexes"
- Nature Communications
- / 2017
- [22]
Yuri Choi, Byungkyun Kang, Jooyong Lee, Sunghu Kim, Gyeong Tae Kim, Hyunju Kang, Bo Ram Lee, Hajin Kim, Sang-Hee Shim, Geunsik Lee, Oh-Hoon Kwon, Byeong-Su Kim
- "Integrative Approach toward Uncovering the Origin of Photoluminescence in Dual Heteroatom-Doped Carbon Nanodots"
- Chemistry of Materials
- / 2016
- [21]
Jejoong Yoo*, Hajin Kim*, Aleksei Aksimentiev+, Taekjip Ha+
- "Direct Evidence for Sequence-Dependent Attraction between Double-Stranded DNA Controlled by Methylation"
- Nature Communications
- / 2016
- [20]
Daniel Sewell, Hajin Kim, Taekjip Ha & Ping Ma
- "A Parameter Estimation Method for Fluorescence Lifetime Data"
- BMC Research Notes
- / 2015
- [19]
Boyang Hua, Kyu Young Han, Ruobo Zhou, Hajin Kim, Xinghua Shi, Sanjaya C Abeysirigunawardena, Ankur Jain, Digvijay Singh, Vasudha Aggarwal, Sarah A Woodson, Taekjip Ha
- "An Improved Surface Passivation Method for Single-Molecule Studies"
- Nature Methods
- / 2014
- [18]
Hajin Kim*, Sanjaya Abeysirigunawardena*, Kaushik Ragunathan, Megan Mayerle, Ke Chen, Zaida Luthey-Schulten, Taekjip Ha+, Sarah Woodson+
- "Protein-Guided RNA Dynamics during Early Ribosome Assembly"
- Nature
- / 2014
- [17]
Jorge A. Lamboy*, Hajin Kim*, Holly Dembinski, Taekjip Ha+, Elizabeth Komives+
- "Single Molecule FRET Reveals the Energy Landscape of the Native State Dynamics of the IkBa Ankyrin Repeat Domain"
- J. Mol. Biol
- / 2013
- [16]
Hajin Kim, Taekjip Ha
- "Single Molecule Nanometry for Biological Physics"
- Reports on Progress in Physics
- / 2013
- [15]
Ibrahim Cisse, Hajin Kim, Taekjip Ha
- "A Rule of Seven in Watson-Crick Base Pairing of Mismatched Sequences"
- Nature Struct. Mol. Biol
- / 2012
- [14]
Ke Chen, John Eargle, Jonathan Lai, Hajin Kim, Taekjip Ha, Megan Mayerle, Sarah Woodson, Zaida Luthey-Schulten
- "Assembly of the Five-way Junction in the Ribosomal Small Subunit Using Hybrid MD-Go Simulations"
- Journal of Physical Chemistry
B
/ 2012- [13]
Hajin Kim, Guo-Qing Tang, Smita S. Patel, Taekjip Ha
- "Opening-Closing Dynamics of the Mitochondrial Transcription Pre-initiation Complex"
- Nucleic Acids Research
(featured article)
/ 2011- [12]
Jorge A. Lamboy*, Hajin Kim*, Kyung Suk Lee, Taekjip Ha+, Elizabeth A. Komives+
- "Visualization of the Nanospring Dynamics of the IκBα Ankyrin Repeat Domain in Real Time"
- Proc. Natl. Acad. Sci
- / 2011
- [11]
Helen Hwang, Hajin Kim, Sua Myong
- "Protein Induced Fluorescence Enhancement as a Single Molecule Assay with Short Distance Sensitivity"
- Proc. Natl. Acad. Sci
- / 2011
- [10]
Byoung-Young Choi, S.-J. Kahng, S. Kim, H. Kim, H. W. Kim, Y. J. Song, J. Ihm & Y. Kuk
- "Conformational Molecular Switch of the Azobenzene Molecule: A Scanning Tunneling Microscopy Study"
- Phys. Rev. Lett
96, 156106
/ 2006- [9]
Sungjun Lee, G. Kim, H. Kim, B.-Y. Choi, J. Lee, B. W. Jeong, J. Ihm, Y. Kuk & S.-J. Kahng
- "Paired Gap States in a Semiconducting Carbon Nanotube: Deep and Shallow Levels"
- Phys. Rev. Lett
95, 166402
/ 2005- [8]
Hajin Kim, J. Lee, S. J. Lee, J.-Y. Park, S.-J. Kahng & Y. Kuk
- "Local Electronic Density of States of a Semiconducting Carbon Nanotube Interface"
- Phys. Rev. B
71, 235402
/ 2005- [7]
Hajin Kim, J, Lee, S. Lee, Y. J. Song, B.-Y. Choi, Y. Kuk & S.-J. Kahng
- "Scanning Tunneling Spectroscopy of a Semiconducting Heterojunction Nanotube on the Au(111) Surface"
- Surf. Sci
581, 241
/ 2005- [6]
Jhinhwan Lee, S. Eggert, H. Kim, S.-J. Kahng, H. Shinohara & Y. Kuk
- "Real Space Imaging of One-Dimensional Standing Waves: Direct Evidence for a Luttinger Liquid"
- Phys. Rev. Lett
93, 166403
/ 2004- [5]
Hajin Kim, J. Lee, Y. J. Song, B. Y. Choi, S.-J. Kahng & Y. Kuk
- "Nano-Scale Structures of a One-Dimensional Junction"
- Thin Solid Films
464, 335
/ 2004- [4]
Hajin Kim, H. J. Chung, J. Lee, S. J. Lee, B.-Y. Choi, Y. J. Song & Y. Kuk
- "Functionalized One-Dimensional Wires and Their Interconnections"
- Jpn. J. Appl. Phys
42, 4780
/ 2003- [3]
Hajin Kim, J. Lee, S.-J. Kahng, Y.-W. Son, S. B. Lee, C.-K. Lee, J. Ihm & Y. Kuk
- "Direct Observation of Localized Defect States in Semiconductor Nanotube Junctions"
- Phys. Rev. Lett
90, 216107
/ 2003- [2]
Hajin Kim, H. J. Chung, J. Lee & Y. Kuk
- "Functionalized One-Dimensional Devices and Their Interconnections"
- J. of Kor. Phys. Soc
42, S134
/ 2003- [1]
Jhinhwan Lee, H. Kim, S.-J. Kahng, G. Kim, Y.-W. Son, J. Ihm, H. Kato, Z. W. Wang, T. Okazaki, H. Shinohara & Y. Kuk
- "Bandgap Modulation of Carbon Nanotubes by Encapsulated Metallofullerenes"
- Nature
415, 1005
/ 2002
[2025]
[2024]
[2023]
[2022]
[2021]
[2020]
[2019]
[2018]
[2017]
[2016]
[2015]
[2014]
[2013]
[2012]
[2011]
[2006]
[2005]
[2004]
[2003]
[2002]
Presentations
- [73] Chanwoo Kim, Jejoong Yoo, Hajin Kim “Periodic Oscillation of Dinucleotide Stiffness as a Dominant Factor in DNA looping”
- 2024 KPS spring Meeting
- 2024-04-26
- PDF File
- [72] Chanwoo Kim, Jejoong Yoo, Hajin Kim “Periodic Oscillation of Dinucleotide Stiffness as a Dominant Factor in DNA looping”
- 2024 KPS
- 2024-01-29
- [71] Chanwoo Kim, Jejoong Yoo, Hajin Kim “Periodic Alteration of DNA Stiffness Generates Intrinsic Chirality in DNA Looping”
- ICBP 2023 International Conference on Biological Physics
- 2024-08-14
- [70] Chanwoo Kim, Yubin Sung, Kei-ichi Takata, Hajin Kim “Conformational Dynamics of Human DNA Polymerase θ during Microhomology-Medicated End Joining”
- 2023 KPS Fall Meeting, October 2023, Changwon Exhibition Convention Center
- 2024-10-24
- [69] Eui-Jin Park, Narendra Chaudhary, Tyler Jepson, Ke Xu, Kyungjae Myung, Hajin Kim, “Super-resolved heterochromatin structure during DNA damage response revealed by CRISPR imaging”
- The 4th Symposium on Biological Physics, Jan 2024, Busan, South Korea
- [68] Jae-Kyeong Im, Brian Choi, Seongmin Yun, Taejun Kwon, Joo H. Kang, Hajin Kim, “Detection of antibiotic resistance gene in single bacterium using in situ Hybridization Chain Reaction”
- International Conference on Advanced Materials and Devices, December 2023, Jeju, South Korea
- [67] Jae-Kyeong Im, Narendra Chaudhary, Eui-Jin Park, Si-Hyeong Nho, Kihyeon Nam, Hajin Kim, “CRISPR RNP Transfection for Live-Cell Imaging of Heterochromatin under DNA Damage Response”
- 2023 KPS Fall Meeting, October 2023, Changwon, South Korea
- [66] Eui-Jin Park, Narendra Chaudhary, Tyler Jepson, Ke Xu, Kyungjae Myung, Hajin Kim, “Super-resolved heterochromatin structure during DNA damage response revealed by CRISPR imaging”
- 2023 KPS Fall Meeting, October 2023, Changwon, South Korea
- [65] Lee Seung-Won, Sohn Byeong-Kwon, Basu Urmimala, Shen Jiayu, Patel Smita, Kim Hajin “Mitochondrial Transcription Dynamics Revealed by Single Molecule FRET Measurement”
- 2023 KPS Fall Meeting, October 2023, Changwon Exhibition Convention Center
- [64] Jae-Kyeong Im, Narendra Chaudhary, Euijin Park, Si-Hyeong Nho, Kihyeon Nam, Hajin Kim* “CRISPR RNP Transfection for Imaging Heterochromatin under DNA Damage Response”
- International Conference on Biological Physics, August 2023, Seoul, South Korea
- [63] Sihyeong Nho, Soyeong An, Seung Min Ahn, Seoyoon Kim, Subin Bae, Hongsoo Lee, Byeongkwon Sohn, Chanho Park, Kipom Kim, Jungmin Kee, Jayil Lee, Duyoung Min, Hajin Kim “Probing Inter-Nucleosomal Interaction with Magnetic Tweezers”
- The 3rd Symposium on Biological Physics, January 2023, Jeju, South Korea
- [62] Eui-Jin Park, Narendra Chaudhary, Tyler Jepson, Minsu Kang, Jae-Kyeong Im, Si-Hyeong Nho, Kihyeon Nam, Jae Sun Ra, Ahmet Yildiz, Kyungjae Myung, Sang-Hee Shim , Ke Xu, Hajin Kim, “Super-resolved heterochromatin structure during DNA damage response revealed by CRISPR imaging”
- BPS (Biophysical Society), February 2023, San Diego, USA
- [61] Seung-Won Lee, Byeong-Kwon Sohn, Urmimala Basu, Hayoon Cho, Jiayu Shen, Aishwarya Deshpande, Smita S. Patel, Hajin Kim “Conformational Dynamics of Mitochondrial Transcription Complex by Single Molecule FRET Measurements”
- The 3rd Symposium on Biological Physics, January 2023, Jeju, South Korea
- [60] Seung-Won Lee, Byeong-Kwon Sohn, Urmimala Basu, Hayoon Cho, Jiayu Shen, Aishwarya Deshpande, Smita S. Patel, Hajin Kim “Initiation-Elongation Transition Dynamics in Yeast and Human Mitochondrial Transcription by Single Molecule FRET Measurements”
- Gordon Research Conferences : Single Molecule Approaches to Biology, July 2022, Castelldefels, B, Spain
- [59] Chanwoo Kim, Yubin Sung, Kei-ichi Takata, Hajin Kim “Mechanism of microhomology mediated end joining by pol theta revealed by single molecule measurements”
- The 12th International Conference on Advanced Materials and Devices, December 2021, Online, South Korea
- [58] Seung-Won Lee, Byeong-Kwon Sohn, Urmimala Basu, Hayoon Cho, Jiayu Shen, Aishwarya Deshpande, Smita S. Patel, Hajin Kim “Conformational Dynamics During Initiation-Elongation Transition in Yeast and Human Mitochondrial Transcription by Single Molecule FRET Measurements”
- International Conference on Advanced Materials and Devices, December 2021, Jeju, South Korea
- [57] Chanwoo Kim, Yubin Sung, Kei-ichi Takata, Hajin Kim “Mechanistic basis for human DNA polymerase θ-directed microhomology-mediated end-joining”
- 2021 KPS Fall Meeting, online South Korea
- [56] Seung-Won Lee, Byeong-Kwon Sohn, Urmimala Basu, Hayoon Cho, Jiayu Shen, Aishwarya Deshpande, Smita S Patel, Hajin Kim, “Single-Molecule Study Reveals the Conformational Dynamics During Initiation-Elongation Transition in Mitochondrial Transcription”
- Biophysical Society, February 2020, San Diego, USA
- [55] Seung-Won Lee, Eunjin Ryu, Sukhyun Kang, and Hajin Kim, “Single Molecule Measurements Reveal Conformational Transitions During DNA Clamp Loading and Unloading”
- Biophysical Society, March 2019, Baltimore, USA
- [54] Seung-Won Lee, Eunjin Ryu, Sukhyun Kang, and Hajin Kim, “Single Molecule Study of ATAD5-Induced Unloading of PCNA”
- Biophysical Society, Jan 2018, San Francisco, USA
- [53] Sihyeong Nho, Hongsoo Lee, Byeong-Kwon Sohn, Chanho Park, Kipom Kim, and Hajin Kim, “Single Molecule Mechanical Measurement of Inter-Nucleosome Interaction”
- The 29th Korean Society for Molecular and Cellular Biology, Jan 2018, Hongcheon, South Korea
- [52] Chanwoo Kim, Jejoong Yoo, Aleksei Aksimentiev, and Hajin Kim, “Single Molecular Study of Looping and Flexibility of Bacterial Genes”
- Multiscale Modeling of Chromatin: Bridging Experiment with Theory, Biophysical Society, April 2019, Les Houches, France
- [51] Chanwoo Kim, Seung-Won Lee, Jejoong Yoo, and Hajin Kim, “Single Molecular Study of Looping and Flexibility of Bacterial Genes”
- The 30th Korean Society for Molecular and Cellular Biology, Jan 2019, Yongpyong, South Korea
- [50] Narendra Chaudhary, Jiwoong Kwon, Sihyeong Nho, Ki-Hyeon Nam, Sang-Hee Shim, and Hajin Kim, “Live cell visualization of chromosome dynamics and super-resolved structure”
- 3D Genome: Gene Regulation and Disease, Keystone Symposia, March 2019, Banff, Canada
- [49] Hyewon Cho, Kawasaki Ryosuke, Shin-ichi Tate, and Hajin Kim, “Exploring the interplay between the protein domains tethered by intrinsically disordered region”
- The 28th Korean Society for Molecular and Cellular Biology (KSMCB), Jan 2017, Yongpyong, South Korea
- [48] Byeong-Kwon Sohn, Hongsoo Lee, Chanho Park, and Hajin Kim, “Measuring the mechanical properties of chromatin with epigenetic modifications using magnetic tweezers”
- The 28th Korean Society for Molecular and Cellular Biology (KSMCB), Jan 2017, Yongpyong, South Korea
- [47] Hayoon Cho, Aishwarya Deshpande, Smita S. patel, and Hajin Kim “Single molecule studies of the transcription initiation dynamics of yeast mitochondria”
- The Korean Society for Molecular and Cellular Biology (KSMCB), Jan 2017, Yongpyong, South Korea
- [46] Narendra Chaudhary, Narangerel Gantumur, Hayoon Cho, Hajin Kim “Multiplexed CRISPR imaging for visualization of real-time chromatin dynamics”
- Chromatin Structure & Function, Gordon Research Conference, May 2016, Les Diablerets, Switzerland
- [45] Narendra Chaudhary, Hayoon Cho, Narangerel Gantumur, Hajin Kim “Imaging and Tracing Multiple Genetic Elements via Multiplexed CRISPR Imaging”
- 60th Biophysical Society Meeting, Mar 2016, Los Angeles http://dx.doi.org/10.1016/j.bpj.2015.11.2181
- [44] Biophysical Mechanism of Sequence-Dependent Attraction Between Double-Stranded DNAs and its Biological Significance
- 60th Biophysical Society Meeting, Mar 2016, Los Angeles
- [43] “Exploring the physical determinant of chromosome structure”
- The 9th International Conference on Advanced Materials and Devices, Dec 2015, Jeju
- [42] Narendra Chaudhary , Min Jong Kim, Narangerel Gantumur, Hayoon Cho, Hajin Kim “Imaging and tracing multiple genetic elements via multiplexed CRISPR imaging”
- The 9th International Conference on Advanced Materials and Devices, Dec 2015, Jeju
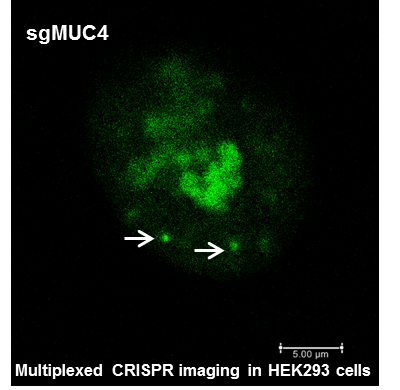
- [41] Hyunju Kang, Jejoong Yoo, Aleksei Aksimentiev, Hajin Kim “Physical mechanism of non-Watson-Crick sequence recognition between double stranded DNA molecules and the role of DNA methylation”
- The 9th International Conference on Advanced Materials and Devices, Dec 2015, Jeju
- [40] Hayoon Cho, Aishwarya Deshpande, Smita S. Patel, Hajin Kim “Single molecule studies on the dynamics of the transcription initiation complex of yeast mitochondria”
- The 9th International Conference on Advanced Materials and Devices, Dec 2015, Jeju
- [39] ByeongKwon Sohn, Hongsoo Lee, Hajin Kim “The role of histone tails in inter-nucleosome interaction and chromatin epigenetics”
- The 9th International Conference on Advanced Materials and Devices, Dec 2015, Jeju
- [38] Hongsoo Lee, Narendra Chaudhary, Hajin Kim “Mapping chromosome territories and intra-chromosomal contacts with DNA fluorescence in situ hybridization”
- The 9th International Conference on Advanced Materials and Devices, Dec 2015, Jeju
- [37] “Exploring the physical determinant of chromosome structure”
- Korean Physical Society Meeting, Oct 2015, Gyeongju
- [36] Narendra Chaudhary, Hayoon Cho, Hajin Kim “Visualizing chromatin dynamics and gene expression in human cells using CRISPR system”
- IUPAC, 45th World Chemistry Congress, Aug 2015, Busan
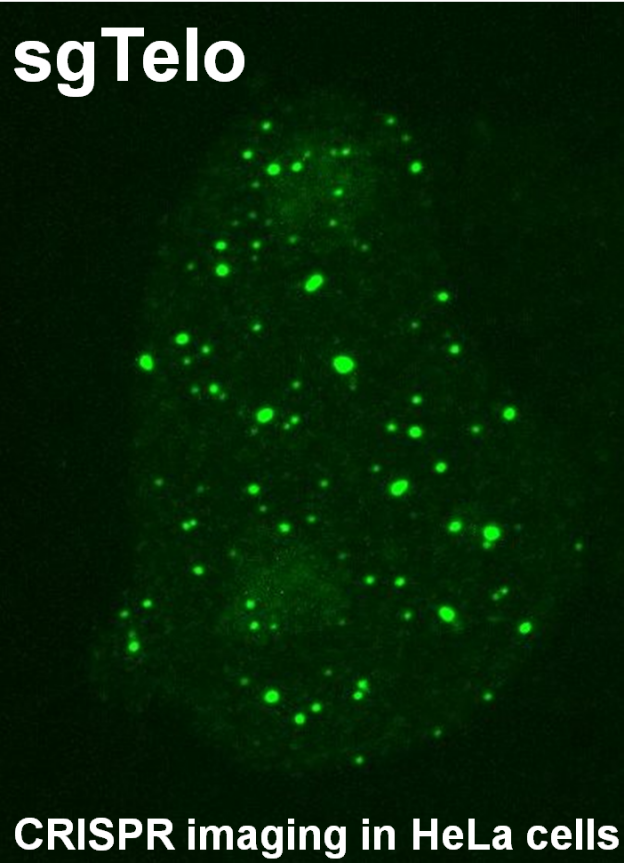
- [35] “DNA sequence composition as a determinant of the chromosome structure”
- Colloquium at Konkuk University, May 2015, Seoul
- [34] “DNA sequence composition as a determinant of the chromosome structure”
- Korean Chemical Society Meeting, April 2015, Seoul
- [33] “Protein-guided RNA dynamics during early ribosome assembly”
- 3rd international symposium on dynamical ordering of biomolecular systems for creation of integrated functions, Jan 2015, Japan
- [32] “Chromosome structure and dynamics and their epigenetic implications”
- Colloquium at POSTECH, Dec 2014, Pohang
- [31] “DNA sequence composition as a determinant of the chromosome structure”
- Colloquium at KAIST, Oct 2014, Daejeon
- [30] “Implications of chromosome structure and dynamics”
- Seminar at KRIBB, Nov 2014, Daejeon
- [29] “DNA sequence composition as a determinant of the chromosome structure”
- Colloquium at Seoul National University, Oct 2014, Seoul
- [28] “Protein-guided RNA dynamics during early ribosome assembly”
- Korean Biophysical Society Symposium, Aug 2014, Sungkyunkwan University
- [27] “AT Content as a Determinant of the Chromosome Structure”
- Gordon Research Conference (Single Molecule Approaches to Biology), Jul 2014, Lucca, Italy
- [26] “From nucleotides to the chromosome: determinant of the chromosome structure and dynamics”
- APCTP Summery School, Jul 2014, Pohang
- [25] “Protein-Guided RNA Dynamics during Early Ribosome Assembly”
- 4th Asia Pacific Protein Association Conference, May 2014, Jeju Island
- [24] “DNA sequence composition as a global determinant of the chromosome structure”
- Colloquium at Sungkyunkwan University, May 2014, Suwon
- [23] “DNA sequence composition as a global determinant of the chromosome structure”
- KPS March Meeting, Apr 2014, Daejun
- [22] “DNA sequence composition as a global determinant of the chromosome structure”
- Colloquium at Korea University, Apr 2014, Seoul
- [21] “AT Content Rules the Chromosome Structure”
- 58th Biophysical Society Meeting, Feb 2014, San Francisco
- [20] “Role of Fluctuations in Biology”
- Seminar at UNIST, Jul 2013, Ulsan
- [19] “Role of Fluctuations in Transcription Initiation and Ribosome Assembly”
- Seminar at Oregon State University, Feb 2013, Corvalis
- [18] “Dynamics of the Mitochondrial Transcription Pre-Initiation Complex”
- EMBO/EMBL Symposium: “The Complex Life of mRNA”, Oct 2012, Heidelberg
- [17] “Protein-Guided RNA Dynamics during Early Ribosome Assembly”
- CPLC Symposium, Sep 2012, UIUC
- [16] “Visualizing Induced Fit during the Early Stage of Ribosome Assembly”
- Gordon Research Conference (Single Molecule Approaches to Biology), July 2012, West Dover
- [15] “Single Molecule Views of Ribosome Assembly”
- 56th Biophysical Society Meeting, Mar 2012, San Diego
- [14] “Role of Fluctuations in Gene Expression and Regulation”
- Seminar at Samsung Advanced Institute of Technology, Feb 2012, Yongin
- [13] “Role of Fluctuations in Biology”
- Seminar at KIAS and Yonsei University (Nano-Medical NCRC), Sep 2011, Seoul
- [12] “Ribosome Assembly in Single Molecule”
- RNA Group Seminar, Aug 2011, UIUC
- [11] “Opening-Closing Dynamics of Transcription Initiation Complex”
- 55th Biophysical Society Meeting, Mar 2011, Baltimore
- [10] “Promoter Opening-Closing Dynamics by Mitochondrial RNA Polymerase”
- 24th Gibbs Conference on Biothermodynamics, Sep 2010, Carbondale
- [9] “IkBa Fluctuation Explored by Single Molecule FRET”
- Mid-West Single-Molecule Workshop, Jul 2010, Washington University
- [8] “GTPase Activation by the Ribosome and the Dynamics of L7/L12”
- UIUC Seminar (CPLC), Feb 2009, Urbana-Champaign
- [7] “Single Porphyrin Molecules as Information Storage Elements”
- 52th American Vacuum Society Meeting, Nov 2005, Boston
- [6] “Luttinger Liquid Behavior of Metallic Carbon Nanotubes Observed in Real Space”
- 13th International Conference on STM/STS and Related Techniques (STM’05), Jul 2005, Sapporo
- [5] “Real Space Imaging of One-Dimensional Standing Waves: Direct Evidence of a Luttinger Liquid”
- Gordon Research Conference on Condensed Matter Physics, Jun 2005, Connecticut College
- [4] “Direct Observation of Localized Defect States in Semiconductor Nanotube Junctions”
- 24th Korean Vacuum Society Meeting, Feb 2003, Hanyang University, Korea
- [3] “Electronic Structure of Carbon Nanotube Intramolecular Junctions”
- Nanotube 2002 Conference, Jul 2002, Boston College
- [2] “Electronic States at the Junctions of Single Wall Carbon Nanotubes”
- 21st European Conference on Surface Science/Nano-7, Jun 2002, Malmö, Sweden
- [1] “Local Bandgap Modulation of Carbon Nanotubes by Encapsulated Metallofullerenes”
- Korean Physical Society Meeting, Oct 2001, Chonnam University, Korea`